rongsheng jin lab made in china

Lam, K. H., Guo, Z., Krez, N., Matsui, T., Perry, K., Weisemann, J., Rummel, A., Bowen, M. E. & Jin, R. A viral-fusion-peptide-like molecular switch drives membrane insertion of botulinum neurotoxin A1. Nat Commun 9, 5367 (2018) doi: 10.1038/s41467-018-07789-4.
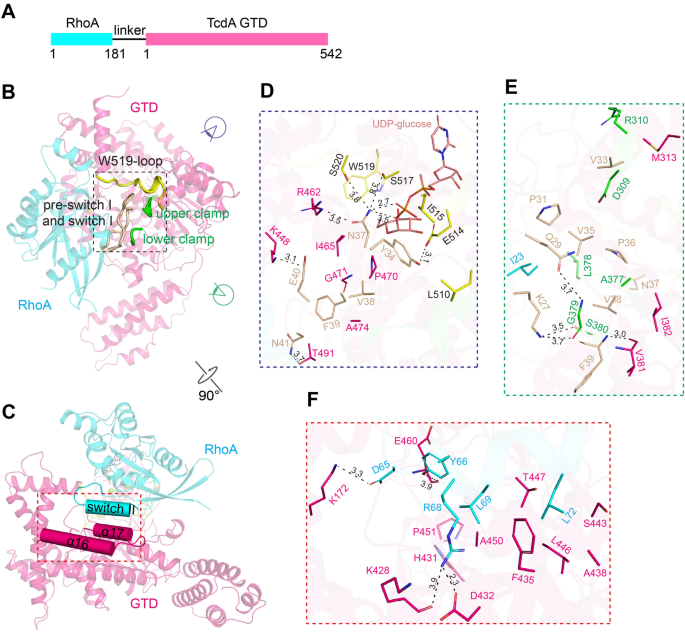
Chen, P., Tao, L., Liu, Z., Dong, M. & Jin, R. Structural insight into Wnt signaling inhibition by Clostridium difficile toxin B. FEBS J (2018) doi: 10.1111/febs.14681.
Chen, P., Tao, L., Wang, T., Zhang, J., He, A., Lam, K. H., Liu, Z., He, X., Perry, K., Dong, M*. & Jin, R*. Structural basis for recognition of frizzled proteins by Clostridium difficile toxin B. Science 360, 664-669 (2018) (*corresponding authors) doi: 10.1126/science.aar1999. PMCID: PMC6231499
Lam, K. H., Sikorra, S., Weisemann, J., Maatsch, H., Perry, K., Rummel, A., Binz, T. & Jin, R. Structural and biochemical characterization of the protease domain of the mosaic botulinum neurotoxin type HA. Pathog Dis 76 (2018) doi: 10.1093/femspd/fty044. PMCID: PMC5961070
Silva, D. A., Stewart, L., Lam, K. H., Jin, R. & Baker, D. Structures and disulfide cross-linking of de novo designed therapeutic mini-proteins. FEBS J 285, 1783-1785 (2018) doi: 10.1111/febs.14394. PMCID: PMC6001749
Lam, K. H., Qi, R., Liu, S., Kroh, A., Yao, G., Perry, K., Rummel, A. & Jin, R. The hypothetical protein P47 of Clostridium botulinum E1 strain Beluga has a structural topology similar to bactericidal/permeability-increasing protein. Toxicon 147, 19-26 (2018) doi: 10.1016/j.toxicon.2017.10.012. PMCID: PMC5902665
Chevalier, A., Silva, D.A., Rocklin, G.J., Hicks, D.R., Vergara, R., Murapa, P., Bernard, S.M., Zhang, L., Lam, K.H., Yao, G., Bahl, C.D., Miyashita, S.I., Goreshnik, I., Fuller, J.T., Koday, M.T., Jenkins, C.M., Colvin, T., Carter, L., Bohn, A., Bryan, C.M., Fernández-Velasco, D.A., Stewart, L., Dong, M., Huang, X., Jin, R., Wilson, I.A., Fuller, D.H. & Baker, D. Massively parallel de novo protein design for targeted therapeutics. Nature 550(7674):74-79 (2017) doi: 10.1038/nature23912. PMCID: PMC5802399
Yao, G., Lam, K.H., Weisemann, J., Peng, L., Krez, N., Perry, K., Shoemaker, C.B., Dong, M., Rummel, A. & Jin, R. A camelid single-domain antibody neutralizes botulinum neurotoxin A by blocking host receptor binding. Sci Rep. 7;7(1):7438. (2017) doi: 10.1038/s41598-017-07457-5. PMCID: PMC5547058
Yao, G., Lam, K.H., Perry, K., Weisemann, J., Rummel, A. & Jin, R. Crystal Structure of the Receptor-Binding Domain of Botulinum Neurotoxin Type HA, Also Known as Type FA or H. Toxins (Basel) 9, 93 (2017) doi: 10.3390/toxins9030093. PMCID: PMC5371848
Yao, G., Zhang, S., Mahrhold, S., Lam, K. H., Stern, D., Bagramyan, K., Perry, K., Kalkum, M., Rummel, A.*, Dong, M.* & Jin, R.* N-linked glycosylation of SV2 is required for binding and uptake of botulinum neurotoxin A. Nat Struct Mol Biol 23 (7):656-662 (2016) (*corresponding authors) doi: 10.1038/nsmb.3245. PMCID: PMC5033645
Lee, K., Lam, K. H., Kruel, A. M., Mahrhold, S., Perry, K., Cheng, L. W., Rummel, A. & Jin, R. Inhibiting oral intoxication of botulinum neurotoxin A complex by carbohydrate receptor mimics. Toxicon 107, 43-49 (2015) doi: 10.1016/j.toxicon.2015.08.003. PMCID: PMC4658216
Lam, K.H. & Jin, R. Architecture of the botulinum neurotoxin complex: a molecular machine for protection and delivery. Current Opinion in Structural Biology 31:89-95 (2015) doi: 10.1016/j.sbi.2015.03.013. PMCID: PMC4476938
Lam, K.H., Yao, G. & Jin, R. Diverse binding modes, same goal: The receptor recognition mechanism of botulinum neurotoxin. Progress in Biophysics and Molecular Biology 117(2-3):225-31 (2015) doi: 10.1016/j.pbiomolbio.2015.02.004. PMCID: PMC4417461
Lam, T.I., Stanker, L.H., Lee, K., Jin, R. & Cheng, L.W. Translocation of botulinum neurotoxin serotype A and associated proteins across the intestinal epithelia. Cellular Microbiology 17(8):1133-1143 (2015) doi: 10.1111/cmi.12424. PMCID: PMC4610714
Matsui, T.*, Gu, S., Lam, K.H., Carter, L.G., Rummel, A., Mathews, II. & Jin, R.* Structural Basis of the pH-Dependent Assembly of a Botulinum Neurotoxin Complex. J. Mol. Biol. 426(22):3773-3782 (2014) doi: 10.1016/j.jmb.2014.09.009. (*corresponding authors) PMCID: PMC4252799
Lee, K., Zhong, X., Gu, S., Kruel, A.M., Dorner, M.B., Perry, K., Rummel, A., Dong, M. & Jin, R. Molecular basis for disruption of E-cadherin adhesion by botulinum neurotoxin A complex. Science 344(6190):1405-1410 (2014) doi: 10.1126/science.1253823. PMCID: PMC4164303
Lee, K., Lam, K.H., Kruel, A.M., Perry, K., Rummel, A. and Jin, R. High-resolution crystal structure of HA33 of botulinum neurotoxin type B progenitor toxin complex. Biochem. Biophys. Res. Commun. 446(2):568-573 (2014) doi: 10.1016/j.bbrc.2014.03.008. PMCID: PMC4020412
Yao, Y., Lee, K., Gu, S., Lam, K.H. & Jin, R. Botulinum Neurotoxin A Complex Recognizes Host Carbohydrates through Its Hemagglutinin Component, Toxins (Basel) 6(2):624-635 (2014) doi: 10.3390/toxins6020624. PMCID: PMC3942755
Lee, K., Gu, S., Jin, L., Le, T.T.N., Cheng, L.W., Strotmeier, J., Kruel, A.M., Yao, G., Perry, K., Rummel, A.* & Jin, R.* Structure of a Bimodular Botulinum Neurotoxin Complex Provides Insights into Its Oral Toxicity. PLoS Pathog. 9(10): e1003690 (2013) doi:10.1371/journal.ppat.1003690. (*corresponding authors) PMCID: PMC3795040
Zong, Y. and Jin, R. Structural mechanisms of the agrin-LRP4-MuSK signaling pathway in neuromuscular junction differentiation. Cell. Mol. Life Sci. 70(17):3077-88 (2013) doi: 10.1007/s00018-012-1209-9. PMCID: PMC4627850
Gu, S. and Jin, R. Assembly and function of the botulinum neurotoxin progenitor complex. Curr. Top. Microbiol. Immunol. 364:21-44 (2013) doi: 10.1007/978-3-642-33570-9_2. PMCID: PMC3875173
Gu, S., Rumpel, S., Zhou, J., Strotmeier, J., Bigalke, H., Perry, K., Shoemaker, C.B., Rummel, A. & Jin, R. Botulinum neurotoxin is shielded by NTNHA in an interlocked complex. Science 335(6071):977-81 (2012) doi: 10.1126/science.1214270. PMCID: PMC3545708
Zong, Y., Zhang, B., Gu, S., Lee, K., Zhou, J., Yao, G., Figueiredo, D., Perry, K., Mei, L.* & Jin, R.* Structural basis of neuron-specific regulation of postsynaptic differentiation. Gene & Development 26:247-258 (2012) doi: 10.1101/gad.180885.111. (*corresponding authors) PMCID: PMC3278892
Yao, G., Zong, Y., Gu, S., Zhou, J., Xu, H., Mathews, II. & Jin, R. Crystal structure of the glutamate receptor GluA1 amino-terminal domain. Biochem. J. 438(2):255-63 (2011) doi: 10.1042/BJ20110801. PMCID: PMC3296483
Strotmeier, J., Gu, S., Jutzi, S., Mahrhold, S., Zhou, J., Pich, A., Eichner, T., Bigalke, H., Rummel, A.*, Jin, R.* & Binz, T*. The biological activity of botulinum neurotoxin type C is dependent upon novel types of ganglioside binding sites. Mol. Microbiol. 81(1):143-56 (2011) doi: 10.1111/j.1365-2958.2011.07682.x. Epub 2011 Jun 2. (*corresponding authors)
Strotmeier, J., Lee, K., Völker, A.K., Mahrhold, S., Zong, Y., Zeiser, J., Zhou, J., Pich, A., Bigalke, H., Binz, T., Rummel, A.* & Jin, R.* Botulinum neurotoxin serotype D attacks neurons via two carbohydrate-binding sites in a ganglioside-dependent manner. Biochem. J. 431(2):207-16 (2010) (*corresponding authors)
Jin, R.*, Singh, S.K., Gu, S., Furukawa, H., Sobolevsky, A.I., Zhou, J., Jin, Y. & Gouaux E.* Crystal structure and association behavior of the GluR2 amino-terminal domain. EMBO J. 28(12):1812-23 (2009) (*corresponding authors) PMCID: PMC2699365
Kumar, J., Schuck. P., Jin, R. & Mayer, M.L. The N-terminal domain of GluR6-subtype glutamate receptor ion channels. Nat. Struct. Mol. Biol. 16(6):631-8 (2009) PMCID: PMC2729365
Jin, R., Rummel, A., Binz, T. & Brunger, A.T. Botulinum neurotoxin B recognizes its protein receptor with high affinity and specificity. Nature 444:1092-5 (2006)
Jin, R., Clark, S., Weeks, A.M., Dudman, J.T., Gouaux, E. & Partin, K.M. Mechanism of positive allosteric modulators acting on AMPA receptors. J. Neurosci. 25(39):9027-36 (2005)
Jin, R., Junutula, J.R., Matern, H.T., Ervin, K.E., Scheller, R.H. & Brunger, A.T. Exo84 and Sec5 are competitive regulatory Sec6/8 effectors to the RalA GTPase. EMBO J. 24:2064-74 (2005)
Jin, R., Bank, T., Mayer, M. L., Traynelis, S. & Gouaux, E. Structural basis for partial agonist action at ionotropic glutamate receptors. Nat. Neurosci. 6(8):803-10 (2003)

Jahid S, Ortega JA, Vuong LM, Acquistapace IM, Hachey SJ, Flesher JL, La Serra MA, Brindani N, La Sala G, Manigrasso J, Arencibia JM, Bertozzi SM, Summa M, Bertorelli R, Armirotti A, Jin R, Liu Z, Chen CF, Edwards R, Hughes CCW, De Vivo M, Ganesan AK. PMID: 35385746; PMCID: PMC9127750.
Chen P, Zeng J, Liu Z, Thaker H, Wang S, Tian S, Zhang J, Tao L, Gutierrez CB, Xing L, Gerhard R, Huang L, Dong M, Jin R. PMID: 34145250; PMCID: PMC8213806.
Chen P, Lam KH, Liu Z, Mindlin FA, Chen B, Gutierrez CB, Huang L, Zhang Y, Hamza T, Feng H, Matsui T, Bowen ME, Perry K, Jin R. PMID: 31308519; PMCID: PMC6684407.

Wild type and mutated recombinant full-length activated BoNT/A1 were produced under biosafety level 2 containment (project number GAA A/Z 40654/3/123) recombinantly in K12 E. coli strain in Dr. Rummel’s lab6-tag were purified on Co2+-Talon matrix (Takara Bio Europe S.A.S., France) and eluted with 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 250 mM imidazole. For proteolytic activation, BoNT/A1 was incubated for 16 h at room temperature with 0.01 U bovine thrombin (Sigma-Aldrich Chemie GmbH, Germany) per µg of BoNT/A1. Subsequent gel filtration (Superdex-200 SEC; GE Healthcare, Germany) was performed in phosphate buffered saline (pH 7.4).
The purified ciA-D12 (S124C) was labeled with Alexa Fluor C2 647 maleimide (Molecular Probes) according to the manufacturer’s instructions. The labeled ciA-D12 was further purified by MonoQ ion-exchange chromatography in 10 mM Hepes (pH 8.0) and eluted with a NaCl gradient. The calculated dye to protein ratio was ~1 mole of dye per mole of ciA-D12. BoNT/A1i-ciA-D12 complex were prepared by mixing BoNT/A1i and ciA-D12 in 1:1.5 molar ratio and the complex was purified by size-exclusion chromatography using Superdex-200.
Liposomes containing 10% GT1b, 69% DOPC, 20% DOPS, 0.5% rhodamine-PE, and 0.5% biotin-PE were prepared by extrusion through a 100 nm pore membrane. To form proteoliposomes, 10 nM BoNT/A1i–D12* or oxidized BoNT/A1iDS–D12* was incubated with 0.5 mg/ml lipid at room temperature for 1 h at the pH indicated. The mixture was then diluted 1000-fold and incubated for 5 minutes in a passivated, quartz microscope chamber functionalized with streptavidin. The biotinylated liposomes were retained and unbound proteins are washed away by exhaustive rinsing with buffer. At the low densities needed for optical resolution of individual liposomes, we could observe sufficient liposomes for statistical analysis, while minimizing the probability that a diffraction-limited spot would contain multiple liposomes. Samples were imaged using a prism-based Total Internal Reflection Fluorescence (TIRF) microscope. Samples were first excited with a laser diode at 640 nm (Newport Corporation, Irvine, CA) to photobleach Alexa 647-labeled BoNT/A1i–D12* or BoNT/A1iDS-D12* molecules followed by excitation with a diode pumped solid state laser at 532 nm (Newport Corporation, Irvine, CA) to probe for Rhodamine-labeled liposomes. Emission from protein and lipids was separated using an Optosplit ratiometric image splitter (Cairn Research Ltd., Faversham UK) containing a 645 nm dichroic mirror, a 585/70 band pass filter for Rhodamine, and a 670/30 band pass filter for Alexa 647 (all Chroma, Bellows Falls, VT). The replicate images were relayed to a single iXon EMCCD camera (Andor Technologies, Belfast, UK) at a frame rate of 10 Hz. Data were processed in MATLAB to cross-correlate the replicate images and extract time traces for diffraction-limited spots with intensity above baseline. Single-molecule traces were hand selected based on the exhibition of single-step decays to baseline during 640 illumination and the appearance of Rhodamine emission during 532 illumination.
Botulism is caused when the botulinum neurotoxin (BoNT) inhibits the release of a neurotransmitter. The disease can be caused by eating toxin-contaminated food, but how the BoNT protein survives the digestive tract and reaches the bloodstream has been a mystery. Last year, a group led by Rongsheng Jin of the University of California, Irvine, demonstrated how a protein called nontoxic nonhemagglutinin (NTNHA) binds to and shields BoNT to protect it from digestive proteases. Jin and colleagues have now used electron microscopy and X-ray crystallography to study a complex of BoNT, NTNHA, and three hemagglutinin proteins that play a role in getting BoNT past intestinal cells to the blood (PLoS Pathog. 2013, DOI: 10.1371/journal.ppat.1003690). The researchers find that the 760-kilodalton complex evokes the construction of the Apollo lunar lander, with BoNT and NTNHA on top and the hemagglutinins forming “legs,” which are the parts that interact with intestinal epithelial cells. The legs land on and bind to sugars on the cells, facilitating passage of BoNT. Jin and colleagues find that dosing mice with a monosaccharide can reduce BoNT toxicity, suggesting a way to prevent—but not treat—botulism. The full protein complex could also point to ways to deliver protein drugs orally.
51Department of Pancreatobiliary Surgery, Nanjing Drum Tower Hospital, the Affiliated Hospital of Nanjing University Medical School, Nanjing, Jiangsu, China
51Department of Pancreatobiliary Surgery, Nanjing Drum Tower Hospital, the Affiliated Hospital of Nanjing University Medical School, Nanjing, Jiangsu, China
Gallbladder cancer (GBC), the sixth most common gastrointestinal tract cancer, poses a significant disease burden in China. However, no national representative data are available on the clinical characteristics, treatment and prognosis of GBC in the Chinese population.
The Chinese Research Group of Gallbladder Cancer (CRGGC) study is a multicentre retrospective registry cohort study. Clinically diagnosed patient with GBC will be identified from 1 January 2008 to December, 2019, by reviewing the electronic medical records from 76 tertiary and secondary hospitals across 28 provinces in China. Patients with pathological and radiological diagnoses of malignancy, including cancer in situ, from the gallbladder and cystic duct are eligible, according to the National Comprehensive Cancer Network 2019 guidelines. Patients will be excluded if GBC is the secondary diagnosis in the discharge summary. The demographic characteristics, medical history, physical examination results, surgery information, pathological data, laboratory examination results and radiology reports will be collected in a standardised case report form. By May 2021, approximately 6000 patient with GBC will be included. The clinical follow-up data will be updated until 5 years after the last admission for GBC of each patient. The study aimed (1) to depict the clinical characteristics, including demographics, pathology, treatment and prognosis of patient with GBC in China; (2) to evaluate the adherence to clinical guidelines of GBC and (3) to improve clinical practice for diagnosing and treating GBC and provide references for policy-makers.
The CRGGC study is a multicentre retrospective registry cohort study. The project was launched by the Shanghai Key Laboratory of Biliary Tract Disease Research, with collaborators from 76 tertiary and secondary hospitals across 28 provinces in China (until 8 March 2020; see online supplemental file 1). We will review the electronic medical records (EMRs) of all diagnosed patient with GBC from 1 January 2008 to December 2019, and extract the related clinical and treatment information. The clinical follow-up data will be updated until 5 years after the last admission of each patient with GBC.
The study will include patients diagnosed before December 2019. According to our preliminary estimation, more than 6000 cases will meet our inclusion criteria. We expect to finish data collection by May 2021. After finishing enrolment of a short-term target of 2000 cases, a primary analysis will be performed. The follow-up will be updated until 5 years after the admission of each patient. More centres are expected to participate in the CRGGC study; thus, the collaborator list may be expanded.
The main outcome is the 5-year overall survival (OS). OS is defined as the duration from the date of first diagnosis to the date of death, and it is censored at the date of the last follow-up when the patients are alive. We will also include the following outcomes: progression-free survival (PFS), defined as the duration between the date of first diagnosis and the date of recurrence, and censored at the date of the last follow-up when the patients have no evidence of recurrence; cancer-specific survival defined as the duration between the date of first diagnosis and the date of cancer-caused death, and censored at the date of the last follow-up when the patients are alive or died from other causes; 3-year OS; and 90-day mortality (for patients who undergo surgery), which will be used to indicate perioperative mortality. Clinical follow-up is defined as the routine practice of hospitals of collecting patient data on treatment, tumour recurrence and patient survival, either by outpatient/inpatient records or telephone. We require hospitals to equip such a system and at least one follow-up per year to join our collaboration. Based on these data, we will update patients’ follow-up statuses every 12 months. The data being collected from clinical follow-up will include date of recurrence, date of death, date of last contact, whether reresection is performed if the malignancy is found incidentally, and whether the patient receives adjuvant therapy.
The workflow of data collection and quality control is shown in figure 1. Before data collection, a group of hepatobiliary specialists designed a structured case report form, aiming to delineate features of patients with GBC and answer corresponding clinical questions. The case report form includes the following information: demographic characteristics, medical history, physical examination results, surgery information, pathological data, laboratory examination results and radiology reports. We have compiled a codebook to standardise the definition of each variable. The data centre will be responsible for training doctors to collect data. Data collection will be carried out by using EpiData (V.4.6.0.2, EpiData Association, Denmark).
After data entry and quality control in each centre, the data will be anonymised and transferred to the servers in the data centre. The data centre is located at Shanghai Key Laboratory of Biliary Tract Disease Research, which is equipped with data servers and essential firewall and backup systems. The data centre will be responsible for quality assessment, storage, sharing and analysis of the data. A group of researchers in the data centre will manage the database.
The data manager will assess the quality of the data after transfer to the data centre. The assessment is based on the structure of missing data and a comparison to baseline data. First, we will apply a grading system, where variables are classified into essential, important and normal importance. Based on the proportion of missing values in each category, the entries will be graded as level A, B, C or D in quality. Entries of category D quality will be normally excluded from analysis. Second, outliers and inconsistent data will be identified. Third, we will compare baseline characteristics of the new data to previous data, with indicators including sex ratio, mean age, proportion of tumor, node, and metastasis (TNM) stage, and 5-year OS. We will apply χ2 test, t-test and log-rank test between the two datasets. When a significant difference is found, the data manager will analyse and record suspicious data. The data manager will inquire about the data in question with the data source and ask for confirmation. The desensitised data will be accessible to collaborators after the completion of the database. A research proposal to the CRGGC Scientific Committee will be essential for analysis of the data.
If the malignancy is diagnosed after surgery, further treatment information may not be available (the patient may turn to a second hospital for reresection). Patients in this case will be categorised as ‘simple cholecystectomy performed; further treatment not available’. If reresection is available, its operative reports will be reviewed as previously mentioned.
Laboratory examination results for patients will be collected with the date of examination. Indicators of interest include the following: (1) routine blood tests: white cell count, haemoglobin and platelet count; (2) liver function tests: total bilirubin, direct bilirubin, albumin, alanine aminotransferase, aspartate aminotransferase, alkaline phosphatase and gamma-glutamyl transferase; (3) renal function tests: blood urea nitrogen and creatine; (4) lipid indicators: triglycerides and total cholesterol; (5) inflammation indicators: C reactive protein and lactic dehydrogenase; (6) coagulation indicators: international normalised ratio, prothrombin time, activated partial thromboplastin time and fibrinogen; (7) tumour markers: carcinoembryonic antigen, carbohydrate antigen 19–9, carbohydrate antigen 125 and alpha fetoprotein; and (8) other tests: blood type and hepatitis B test. The test method and normal range of each indicator may vary across hospitals. Thus, we will first uniform the units of each indicator according to the first enrolled hospital; then, based on the first enrolled hospital, we will normalise each result of laboratory examination by its normal range across different hospitals.
On reviewing the published studies on GBC, we found a lack of large observational studies on GBC in China focusing on its clinical features and prognosis. Moreover, international studies on GBC were limited by small sample sizes and inconsistent coding systems for GBC. Our data will establish a collaborative platform for GBC research, providing valuable data from China.
GBC cases in China account for nearly 1/4 of cases worldwide; thus, GBC poses a significant disease burden in China. However, few clinical studies of the diagnosis and treatment of GBC have been performed in China, making this significant population under-represented. By launching the CRGGC study, we also expect to boost collaborations among Chinese researchers. We hope this collaboration could induce further translational research and clinical trials in China, providing essential evidence on GBC treatment.
There are several limitations and potential biases in our study design. (1) The retrospective nature is inevitably related to information bias and heterogeneity in the data recording. This will cause difficulty in the standardisation of data and a relatively large proportion of missing data. To overcome such bias, we composed and continue to update a codebook for standardisation of each variable. Researchers responsible for data entry are trained and qualified at the data centre. The missing data are analysed to determine potential bias. (2) This is a retrospective study using convenience sampling. Thus, the cohort may not be completely representative of patient with GBC in China. However, we attempt to include centres in every province in China. Moreover, most patients with cancer in China are treated in tertiary hospitals. (3) Biospecimens of the involved patients are not collected. Future collaboration on this issue will be considered. (4) As patients with incidental GBC may turn to other hospitals for reresection, resulting in incomplete treatment information. We addressed this problem by defining these patients separately to aid further sensitivity analysis. (5) Currently, we have not made collaboration with Chinese death registry, thus, part of the death information may be lost and the follow-up data might be biased due to lack of validation. On the one hand, the CRGGC study actively seek cooperation with relevant registries; on the other hand, we require collaborated hospitals to equip clinical follow-up system, compare prognosis data in each hospital to identify systematic bias, and update follow-up data yearly.
We would like to thank Professor Wenyi Yang at Shanghai General Hospital, China, and Professor Jiong Li at Aarhus University, Denmark, for their valuable advice on the CRGGC registry. We thank our collaborators for their contributions to the CRGGC study, as follows: Professor Lianxin Liu at Anhui Provincial Hospital, Professor Xu Liu at Peking University Shenzhen Hospital, Professor Yinmo Yang at Peking University First Hospital, Professor Qiang Xu at Peking Union Medical College Hospital, Professor Banghao Xu at The First Affiliated Hospital of Guangxi Medical University, Professor Jianhua Liu at The Second Hospital of Hebei Medical University, Professor Baobiing Yin at Huashan Hospital, Professor Weilong Cai at Huzhou Central Hospital, Professor Zhiping Zhang at Ningbo First Hospital, Professor Xuting Zhi at Qilu Hospital of Shandong University, Professor Longhua Rao at Central Hospital of Minhang District, Professor Xiaoping Yang at Shanghai Pudong Hospital, Professor Jiahua Yang at Putuo District People’s Hospital of Shanghai, Professor Ruiwu Dai at Chengdu Military General Hospital, Professor Leida Zhang at Southwest Hospital, Professor Xinbao Wang at Zhejiang Cancer Hospital, Professor Jinhui Zhou at The Second Affiliated Hospital of Zhejiang University School of Medicine, Professor Yongjun Chen at Ruijin Hospital, and Professor Yongwei Sun at Renji Hospital. We would also like to thank the researchers who made significant contributions to the CRGGC registry, as follows: Professor Min Wang at Tongji Hospital, Professor Rongsheng Zhang at Shanxi Provincial Cancer Hospital, Dr Kai Qu at The First Affiliated Hospital of Xi"an Jiaotong University, Professor Lei Zou at The First Affiliated Hospital of Kunming Medical University, Professor Fubao Liu at The First Affiliated Hospital of Anhui Medical University, Professor Leibo Xu at Sun Yat-Sen Memorial Hospital, Sun Yat-Sen University, Professor Xianhai Mao at People"s Hospital of Hunan Province, Professor Ling Zhang at Henan Cancer Hospital, Dr Zhizhen Li at Eastern Hepatobiliary Surgery Hospital, and Dr Lei Wang at Wuxi Second People"s Hospital.
Contributors: YL is the principal investigator steering the CRGGC, and responsible for conceptualisation, funding acquisition and supervision of the study. TR, YL, XZ and YG wrote and revised the manuscript.TR, YL, YG, ZS, ML, XW, X-AW, WW, YS, RB, WG and PD discussed and drafted the case report form, standard operation procedure in data collection and management, and standard of quality control. TR and YL are responsible for data curation and coordination.XZ is responsible for the methodology. TR, YL and XZ will be responsible for data analysis and reporting of the work. XD, ChanL, ChangL, BS, JL, LW, DH, RQ, XJ, XZ, JX, JJ, BY, BL, CD, JC, HC, FT, ZZ, YW, HJ, HC, ZF, JG, WH, XF, LF, LZ, CZ, KW, XZ, XL, CJ, YQ, YC, YX, XW, HL, YH, CL, JH, CW, QL, XL, JL, ML, YQ, BW, JZ, XC, HZ, KH, MY, PW, HZ, XM, JH, WG and YL are responsible for resources, data collection and quality control in collaborated hospitals. All authors reviewed the manuscript for intellectual content and approved the final version of the report.
Funding: This study was supported by the National Natural Science Foundation of China (No. 31620103910,81874181, and 91940305), the Emerging Frontier Programme of Hospital Development Centre (No. SHDC12018107), the Key Programme of Shanghai Science and Technology Commission (No. YDZX20193100004049), the Shanghai Key Laboratory of Biliary Tract Disease Research Foundation (17DZ2260200), the Shanghai Artificial Intelligence Innovation and Development Project (2019, Project of Xinhua Hospital Affiliated to Shanghai Jiao Tong University School of Medicine), the Multi-Centre Clinical Research Project of Shanghai Jiao Tong University School of Medicine (DLY201507) and the Translational Medicine Innovation Fund of Shanghai Jiao Tong University School of Medicine (15ZH4003).
7. NCCN clinical practice guidelines in oncology: hepatobiliary cancers, version 4. 2019: national comprehensive cancer network, 2019. Available: https://www.nccn.org/professionals/physician_gls/pdf/hepatobiliary.pdf [Accessed 2020-02-13]. PubMed]
Collaborative stage data collection system user documentation and coding instructions. version 02.04. Chicago, IL: American Joint Committee on Cancer, 2012.

2. The meeting registration is an integral package. We encourage all our participants to stay for the full meeting period and communicate with each other. No refund/discount or day pass is available for partial participation.

abstract = {Mixed-chirality peptide macrocycles such as cyclosporine are among the most potent therapeutics identified to date, but there is currently no way to systematically search the structural space spanned by such compounds. Natural proteins do not provide a useful guide: Peptide macrocycles lack regular secondary structures and hydrophobic cores, and can contain local structures not accessible with L-amino acids. Here, we enumerate the stable structures that can be adopted by macrocyclic peptides composed of L- and D-amino acids by near-exhaustive backbone sampling followed by sequence design and energy landscape calculations. We identify more than 200 designs predicted to fold into single stable structures, many times more than the number of currently available unbound peptide macrocycle structures. Nuclear magnetic resonance structures of 9 of 12 designed 7- to 10-residue macrocycles, and three 11- to 14-residue bicyclic designs, are close to the computational models. Our results provide a nearly complete coverage of the rich space of structures possible for short peptide macrocycles and vastly increase the available starting scaffolds for both rational drug design and library selection methods.},
Mixed-chirality peptide macrocycles such as cyclosporine are among the most potent therapeutics identified to date, but there is currently no way to systematically search the structural space spanned by such compounds. Natural proteins do not provide a useful guide: Peptide macrocycles lack regular secondary structures and hydrophobic cores, and can contain local structures not accessible with L-amino acids. Here, we enumerate the stable structures that can be adopted by macrocyclic peptides composed of L- and D-amino acids by near-exhaustive backbone sampling followed by sequence design and energy landscape calculations. We identify more than 200 designs predicted to fold into single stable structures, many times more than the number of currently available unbound peptide macrocycle structures. Nuclear magnetic resonance structures of 9 of 12 designed 7- to 10-residue macrocycles, and three 11- to 14-residue bicyclic designs, are close to the computational models. Our results provide a nearly complete coverage of the rich space of structures possible for short peptide macrocycles and vastly increase the available starting scaffolds for both rational drug design and library selection methods.
Aaron Chevalier*, Daniel-Adriano Silva*, Gabriel J. Rocklin*, Derrick R. Hicks, Renan Vergara, Patience Murapa, Steffen M. Bernard, Lu Zhang, Kwok-Ho Lam, Guorui Yao, Christopher D. Bahl, Shin-Ichiro Miyashita, Inna Goreshnik, James T. Fuller andMerika T. Koday, Cody M. Jenkins, Tom Colvin, Lauren Carter, Alan Bohn, Cassie M. Bryan, D. Alejandro Fernández-Velasco, Lance Stewart, Min Dong, Xuhui Huang, Rongsheng Jin, Ian A. Wilson, Deborah H. Fuller, David Baker
author = {Aaron Chevalier* and Daniel-Adriano Silva* and Gabriel J. Rocklin* and Derrick R. Hicks and Renan Vergara and Patience Murapa and Steffen M. Bernard and Lu Zhang and Kwok-Ho Lam and Guorui Yao and Christopher D. Bahl and Shin-Ichiro Miyashita and Inna Goreshnik and James T. Fuller andMerika T. Koday and Cody M. Jenkins and Tom Colvin and Lauren Carter and Alan Bohn and Cassie M. Bryan and D. Alejandro Fernández-Velasco and Lance Stewart and Min Dong and Xuhui Huang and Rongsheng Jin and Ian A. Wilson and Deborah H. Fuller and David Baker},
Marcos, Enrique*, Basanta, Benjamin*, Chidyausiku, Tamuka M., Tang, Yuefeng, Oberdorfer, Gustav, Liu, Gaohua, Swapna, G. V. T., Guan, Rongjin, Silva, Daniel-Adriano, Dou, Jiayi, Pereira, Jose Henrique, Xiao, Rong, Sankaran, Banumathi, Zwart, Peter H., Montelione, Gaetano T., Baker, David
author = {Marcos, Enrique* and Basanta, Benjamin* and Chidyausiku, Tamuka M. and Tang, Yuefeng and Oberdorfer, Gustav and Liu, Gaohua and Swapna, G. V. T. and Guan, Rongjin and Silva, Daniel-Adriano and Dou, Jiayi and Pereira, Jose Henrique and Xiao, Rong and Sankaran, Banumathi and Zwart, Peter H. and Montelione, Gaetano T. and Baker, David},
abstract = {Nature provides many examples of self- and co-assembling protein-based molecular machines, including icosahedral protein cages that serve as scaffolds, enzymes, and compartments for essential biochemical reactions and icosahedral virus capsids, which encapsidate and protect viral genomes and mediate entry into host cells. Inspired by these natural materials, we report the computational design and experimental characterization of co-assembling, two-component, 120-subunit icosahedral protein nanostructures with molecular weights (1.8 to 2.8 megadaltons) and dimensions (24 to 40 nanometers in diameter) comparable to those of small viral capsids. Electron microscopy, small-angle x-ray scattering, and x-ray crystallography show that 10 designs spanning three distinct icosahedral architectures form materials closely matching the design models. In vitro assembly of icosahedral complexes from independently purified components occurs rapidly, at rates comparable to those of viral capsids, and enables controlled packaging of molecular cargo through charge complementarity. The ability to design megadalton-scale materials with atomic-level accuracy and controllable assembly opens the door to a new generation of genetically programmable protein-based molecular machines.},
Nature provides many examples of self- and co-assembling protein-based molecular machines, including icosahedral protein cages that serve as scaffolds, enzymes, and compartments for essential biochemical reactions and icosahedral virus capsids, which encapsidate and protect viral genomes and mediate entry into host cells. Inspired by these natural materials, we report the computational design and experimental characterization of co-assembling, two-component, 120-subunit icosahedral protein nanostructures with molecular weights (1.8 to 2.8 megadaltons) and dimensions (24 to 40 nanometers in diameter) comparable to those of small viral capsids. Electron microscopy, small-angle x-ray scattering, and x-ray crystallography show that 10 designs spanning three distinct icosahedral architectures form materials closely matching the design models. In vitro assembly of icosahedral complexes from independently purified components occurs rapidly, at rates comparable to those of viral capsids, and enables controlled packaging of molecular cargo through charge complementarity. The ability to design megadalton-scale materials with atomic-level accuracy and controllable assembly opens the door to a new generation of genetically programmable protein-based molecular machines.




 8613371530291
8613371530291