rongsheng jin lab price

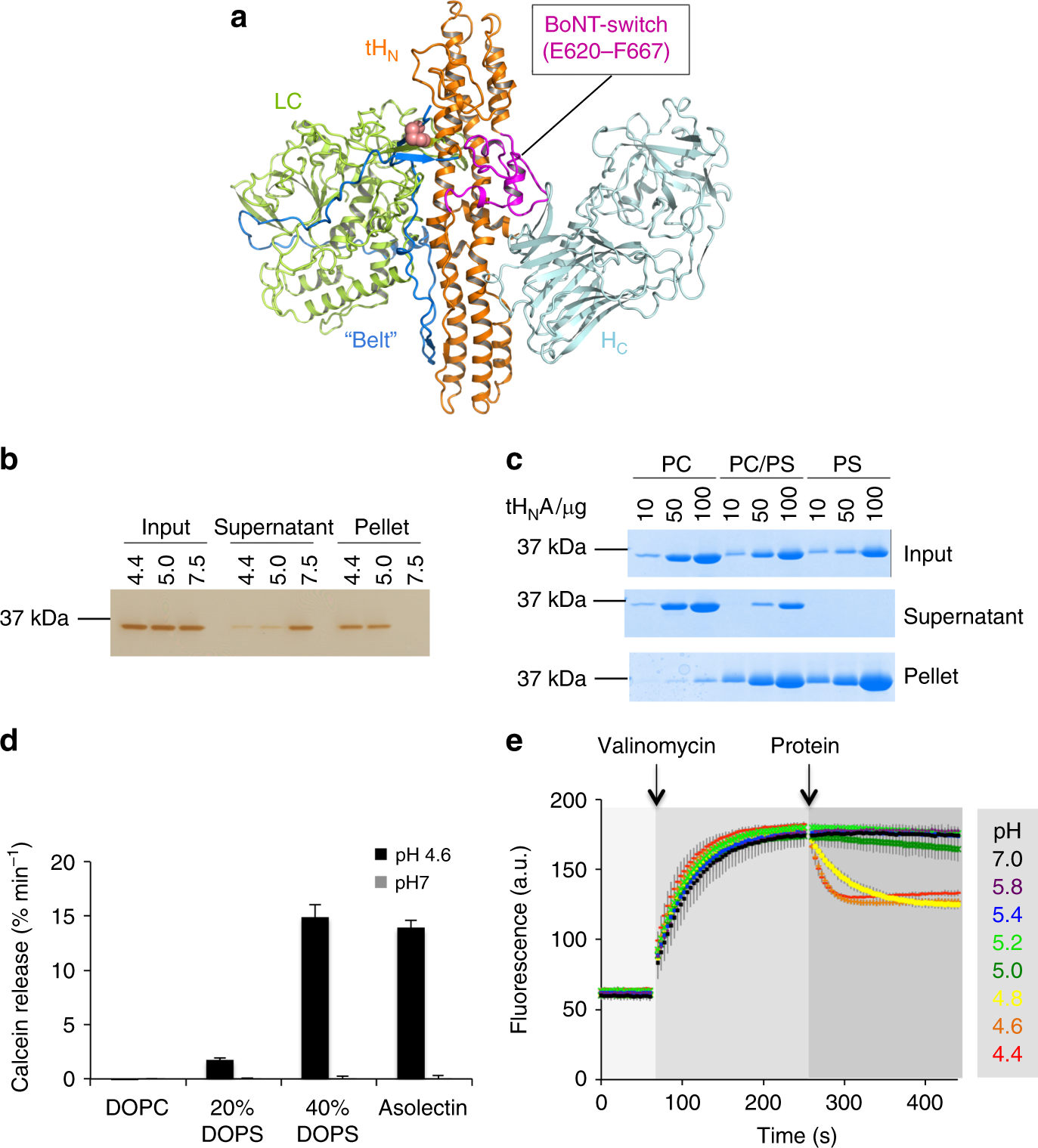
Lam, K. H., Guo, Z., Krez, N., Matsui, T., Perry, K., Weisemann, J., Rummel, A., Bowen, M. E. & Jin, R. A viral-fusion-peptide-like molecular switch drives membrane insertion of botulinum neurotoxin A1. Nat Commun 9, 5367 (2018) doi: 10.1038/s41467-018-07789-4.
Chen, P., Tao, L., Liu, Z., Dong, M. & Jin, R. Structural insight into Wnt signaling inhibition by Clostridium difficile toxin B. FEBS J (2018) doi: 10.1111/febs.14681.
Chen, P., Tao, L., Wang, T., Zhang, J., He, A., Lam, K. H., Liu, Z., He, X., Perry, K., Dong, M*. & Jin, R*. Structural basis for recognition of frizzled proteins by Clostridium difficile toxin B. Science 360, 664-669 (2018) (*corresponding authors) doi: 10.1126/science.aar1999. PMCID: PMC6231499
Lam, K. H., Sikorra, S., Weisemann, J., Maatsch, H., Perry, K., Rummel, A., Binz, T. & Jin, R. Structural and biochemical characterization of the protease domain of the mosaic botulinum neurotoxin type HA. Pathog Dis 76 (2018) doi: 10.1093/femspd/fty044. PMCID: PMC5961070
Silva, D. A., Stewart, L., Lam, K. H., Jin, R. & Baker, D. Structures and disulfide cross-linking of de novo designed therapeutic mini-proteins. FEBS J 285, 1783-1785 (2018) doi: 10.1111/febs.14394. PMCID: PMC6001749
Lam, K. H., Qi, R., Liu, S., Kroh, A., Yao, G., Perry, K., Rummel, A. & Jin, R. The hypothetical protein P47 of Clostridium botulinum E1 strain Beluga has a structural topology similar to bactericidal/permeability-increasing protein. Toxicon 147, 19-26 (2018) doi: 10.1016/j.toxicon.2017.10.012. PMCID: PMC5902665
Chevalier, A., Silva, D.A., Rocklin, G.J., Hicks, D.R., Vergara, R., Murapa, P., Bernard, S.M., Zhang, L., Lam, K.H., Yao, G., Bahl, C.D., Miyashita, S.I., Goreshnik, I., Fuller, J.T., Koday, M.T., Jenkins, C.M., Colvin, T., Carter, L., Bohn, A., Bryan, C.M., Fernández-Velasco, D.A., Stewart, L., Dong, M., Huang, X., Jin, R., Wilson, I.A., Fuller, D.H. & Baker, D. Massively parallel de novo protein design for targeted therapeutics. Nature 550(7674):74-79 (2017) doi: 10.1038/nature23912. PMCID: PMC5802399
Yao, G., Lam, K.H., Weisemann, J., Peng, L., Krez, N., Perry, K., Shoemaker, C.B., Dong, M., Rummel, A. & Jin, R. A camelid single-domain antibody neutralizes botulinum neurotoxin A by blocking host receptor binding. Sci Rep. 7;7(1):7438. (2017) doi: 10.1038/s41598-017-07457-5. PMCID: PMC5547058
Yao, G., Lam, K.H., Perry, K., Weisemann, J., Rummel, A. & Jin, R. Crystal Structure of the Receptor-Binding Domain of Botulinum Neurotoxin Type HA, Also Known as Type FA or H. Toxins (Basel) 9, 93 (2017) doi: 10.3390/toxins9030093. PMCID: PMC5371848
Yao, G., Zhang, S., Mahrhold, S., Lam, K. H., Stern, D., Bagramyan, K., Perry, K., Kalkum, M., Rummel, A.*, Dong, M.* & Jin, R.* N-linked glycosylation of SV2 is required for binding and uptake of botulinum neurotoxin A. Nat Struct Mol Biol 23 (7):656-662 (2016) (*corresponding authors) doi: 10.1038/nsmb.3245. PMCID: PMC5033645
Lee, K., Lam, K. H., Kruel, A. M., Mahrhold, S., Perry, K., Cheng, L. W., Rummel, A. & Jin, R. Inhibiting oral intoxication of botulinum neurotoxin A complex by carbohydrate receptor mimics. Toxicon 107, 43-49 (2015) doi: 10.1016/j.toxicon.2015.08.003. PMCID: PMC4658216
Lam, K.H. & Jin, R. Architecture of the botulinum neurotoxin complex: a molecular machine for protection and delivery. Current Opinion in Structural Biology 31:89-95 (2015) doi: 10.1016/j.sbi.2015.03.013. PMCID: PMC4476938
Lam, K.H., Yao, G. & Jin, R. Diverse binding modes, same goal: The receptor recognition mechanism of botulinum neurotoxin. Progress in Biophysics and Molecular Biology 117(2-3):225-31 (2015) doi: 10.1016/j.pbiomolbio.2015.02.004. PMCID: PMC4417461
Lam, T.I., Stanker, L.H., Lee, K., Jin, R. & Cheng, L.W. Translocation of botulinum neurotoxin serotype A and associated proteins across the intestinal epithelia. Cellular Microbiology 17(8):1133-1143 (2015) doi: 10.1111/cmi.12424. PMCID: PMC4610714
Matsui, T.*, Gu, S., Lam, K.H., Carter, L.G., Rummel, A., Mathews, II. & Jin, R.* Structural Basis of the pH-Dependent Assembly of a Botulinum Neurotoxin Complex. J. Mol. Biol. 426(22):3773-3782 (2014) doi: 10.1016/j.jmb.2014.09.009. (*corresponding authors) PMCID: PMC4252799
Lee, K., Zhong, X., Gu, S., Kruel, A.M., Dorner, M.B., Perry, K., Rummel, A., Dong, M. & Jin, R. Molecular basis for disruption of E-cadherin adhesion by botulinum neurotoxin A complex. Science 344(6190):1405-1410 (2014) doi: 10.1126/science.1253823. PMCID: PMC4164303
Lee, K., Lam, K.H., Kruel, A.M., Perry, K., Rummel, A. and Jin, R. High-resolution crystal structure of HA33 of botulinum neurotoxin type B progenitor toxin complex. Biochem. Biophys. Res. Commun. 446(2):568-573 (2014) doi: 10.1016/j.bbrc.2014.03.008. PMCID: PMC4020412
Yao, Y., Lee, K., Gu, S., Lam, K.H. & Jin, R. Botulinum Neurotoxin A Complex Recognizes Host Carbohydrates through Its Hemagglutinin Component, Toxins (Basel) 6(2):624-635 (2014) doi: 10.3390/toxins6020624. PMCID: PMC3942755
Lee, K., Gu, S., Jin, L., Le, T.T.N., Cheng, L.W., Strotmeier, J., Kruel, A.M., Yao, G., Perry, K., Rummel, A.* & Jin, R.* Structure of a Bimodular Botulinum Neurotoxin Complex Provides Insights into Its Oral Toxicity. PLoS Pathog. 9(10): e1003690 (2013) doi:10.1371/journal.ppat.1003690. (*corresponding authors) PMCID: PMC3795040
Zong, Y. and Jin, R. Structural mechanisms of the agrin-LRP4-MuSK signaling pathway in neuromuscular junction differentiation. Cell. Mol. Life Sci. 70(17):3077-88 (2013) doi: 10.1007/s00018-012-1209-9. PMCID: PMC4627850
Gu, S. and Jin, R. Assembly and function of the botulinum neurotoxin progenitor complex. Curr. Top. Microbiol. Immunol. 364:21-44 (2013) doi: 10.1007/978-3-642-33570-9_2. PMCID: PMC3875173
Gu, S., Rumpel, S., Zhou, J., Strotmeier, J., Bigalke, H., Perry, K., Shoemaker, C.B., Rummel, A. & Jin, R. Botulinum neurotoxin is shielded by NTNHA in an interlocked complex. Science 335(6071):977-81 (2012) doi: 10.1126/science.1214270. PMCID: PMC3545708
Zong, Y., Zhang, B., Gu, S., Lee, K., Zhou, J., Yao, G., Figueiredo, D., Perry, K., Mei, L.* & Jin, R.* Structural basis of neuron-specific regulation of postsynaptic differentiation. Gene & Development 26:247-258 (2012) doi: 10.1101/gad.180885.111. (*corresponding authors) PMCID: PMC3278892
Yao, G., Zong, Y., Gu, S., Zhou, J., Xu, H., Mathews, II. & Jin, R. Crystal structure of the glutamate receptor GluA1 amino-terminal domain. Biochem. J. 438(2):255-63 (2011) doi: 10.1042/BJ20110801. PMCID: PMC3296483
Strotmeier, J., Gu, S., Jutzi, S., Mahrhold, S., Zhou, J., Pich, A., Eichner, T., Bigalke, H., Rummel, A.*, Jin, R.* & Binz, T*. The biological activity of botulinum neurotoxin type C is dependent upon novel types of ganglioside binding sites. Mol. Microbiol. 81(1):143-56 (2011) doi: 10.1111/j.1365-2958.2011.07682.x. Epub 2011 Jun 2. (*corresponding authors)
Strotmeier, J., Lee, K., Völker, A.K., Mahrhold, S., Zong, Y., Zeiser, J., Zhou, J., Pich, A., Bigalke, H., Binz, T., Rummel, A.* & Jin, R.* Botulinum neurotoxin serotype D attacks neurons via two carbohydrate-binding sites in a ganglioside-dependent manner. Biochem. J. 431(2):207-16 (2010) (*corresponding authors)
Jin, R.*, Singh, S.K., Gu, S., Furukawa, H., Sobolevsky, A.I., Zhou, J., Jin, Y. & Gouaux E.* Crystal structure and association behavior of the GluR2 amino-terminal domain. EMBO J. 28(12):1812-23 (2009) (*corresponding authors) PMCID: PMC2699365
Kumar, J., Schuck. P., Jin, R. & Mayer, M.L. The N-terminal domain of GluR6-subtype glutamate receptor ion channels. Nat. Struct. Mol. Biol. 16(6):631-8 (2009) PMCID: PMC2729365
Jin, R., Rummel, A., Binz, T. & Brunger, A.T. Botulinum neurotoxin B recognizes its protein receptor with high affinity and specificity. Nature 444:1092-5 (2006)
Jin, R., Clark, S., Weeks, A.M., Dudman, J.T., Gouaux, E. & Partin, K.M. Mechanism of positive allosteric modulators acting on AMPA receptors. J. Neurosci. 25(39):9027-36 (2005)
Jin, R., Junutula, J.R., Matern, H.T., Ervin, K.E., Scheller, R.H. & Brunger, A.T. Exo84 and Sec5 are competitive regulatory Sec6/8 effectors to the RalA GTPase. EMBO J. 24:2064-74 (2005)
Jin, R., Bank, T., Mayer, M. L., Traynelis, S. & Gouaux, E. Structural basis for partial agonist action at ionotropic glutamate receptors. Nat. Neurosci. 6(8):803-10 (2003)

Jahid S, Ortega JA, Vuong LM, Acquistapace IM, Hachey SJ, Flesher JL, La Serra MA, Brindani N, La Sala G, Manigrasso J, Arencibia JM, Bertozzi SM, Summa M, Bertorelli R, Armirotti A, Jin R, Liu Z, Chen CF, Edwards R, Hughes CCW, De Vivo M, Ganesan AK. PMID: 35385746; PMCID: PMC9127750.
Chen P, Zeng J, Liu Z, Thaker H, Wang S, Tian S, Zhang J, Tao L, Gutierrez CB, Xing L, Gerhard R, Huang L, Dong M, Jin R. PMID: 34145250; PMCID: PMC8213806.
Chen P, Lam KH, Liu Z, Mindlin FA, Chen B, Gutierrez CB, Huang L, Zhang Y, Hamza T, Feng H, Matsui T, Bowen ME, Perry K, Jin R. PMID: 31308519; PMCID: PMC6684407.

Wild type and mutated recombinant full-length activated BoNT/A1 were produced under biosafety level 2 containment (project number GAA A/Z 40654/3/123) recombinantly in K12 E. coli strain in Dr. Rummel’s lab6-tag were purified on Co2+-Talon matrix (Takara Bio Europe S.A.S., France) and eluted with 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 250 mM imidazole. For proteolytic activation, BoNT/A1 was incubated for 16 h at room temperature with 0.01 U bovine thrombin (Sigma-Aldrich Chemie GmbH, Germany) per µg of BoNT/A1. Subsequent gel filtration (Superdex-200 SEC; GE Healthcare, Germany) was performed in phosphate buffered saline (pH 7.4).
The purified ciA-D12 (S124C) was labeled with Alexa Fluor C2 647 maleimide (Molecular Probes) according to the manufacturer’s instructions. The labeled ciA-D12 was further purified by MonoQ ion-exchange chromatography in 10 mM Hepes (pH 8.0) and eluted with a NaCl gradient. The calculated dye to protein ratio was ~1 mole of dye per mole of ciA-D12. BoNT/A1i-ciA-D12 complex were prepared by mixing BoNT/A1i and ciA-D12 in 1:1.5 molar ratio and the complex was purified by size-exclusion chromatography using Superdex-200.
Liposomes containing 10% GT1b, 69% DOPC, 20% DOPS, 0.5% rhodamine-PE, and 0.5% biotin-PE were prepared by extrusion through a 100 nm pore membrane. To form proteoliposomes, 10 nM BoNT/A1i–D12* or oxidized BoNT/A1iDS–D12* was incubated with 0.5 mg/ml lipid at room temperature for 1 h at the pH indicated. The mixture was then diluted 1000-fold and incubated for 5 minutes in a passivated, quartz microscope chamber functionalized with streptavidin. The biotinylated liposomes were retained and unbound proteins are washed away by exhaustive rinsing with buffer. At the low densities needed for optical resolution of individual liposomes, we could observe sufficient liposomes for statistical analysis, while minimizing the probability that a diffraction-limited spot would contain multiple liposomes. Samples were imaged using a prism-based Total Internal Reflection Fluorescence (TIRF) microscope. Samples were first excited with a laser diode at 640 nm (Newport Corporation, Irvine, CA) to photobleach Alexa 647-labeled BoNT/A1i–D12* or BoNT/A1iDS-D12* molecules followed by excitation with a diode pumped solid state laser at 532 nm (Newport Corporation, Irvine, CA) to probe for Rhodamine-labeled liposomes. Emission from protein and lipids was separated using an Optosplit ratiometric image splitter (Cairn Research Ltd., Faversham UK) containing a 645 nm dichroic mirror, a 585/70 band pass filter for Rhodamine, and a 670/30 band pass filter for Alexa 647 (all Chroma, Bellows Falls, VT). The replicate images were relayed to a single iXon EMCCD camera (Andor Technologies, Belfast, UK) at a frame rate of 10 Hz. Data were processed in MATLAB to cross-correlate the replicate images and extract time traces for diffraction-limited spots with intensity above baseline. Single-molecule traces were hand selected based on the exhibition of single-step decays to baseline during 640 illumination and the appearance of Rhodamine emission during 532 illumination.

GUID: 97AA2E70-9FB0-4537-BD4C-0598D4570100Whole genome sequencing data are available in the National Center for Biotechnology Information database under the BioProject PRJNA751216.
The constructed surrogate strain C. botulinum Beluga Ei carries unique restriction digestion sites as molecular markers to confirm the presence of the inactivating mutations and to facilitate identification. Being a genetically modified organism, C. botulinum Beluga Ei is intended exclusively for use in contained research facilities. Even in the very unlikely event of accidental release, potentially leading to Beluga Ei being erroneously detected as pathogenic C. botulinum strain when applying standard serological and DNA-based detection methodsC. botulinum should carry toxoid-specific molecular markers and that they must be handled only in the specialized research laboratories. In conclusion, the constructed C. botulinum Beluga Ei strain producing biologically inactive BoNT/Ei provides a safe and stable surrogate for botulinum neurotoxin research and for food safety risk assessment.
The C. botulinum Beluga Ei strain has been officially excluded from the US select agent regulations of the Division of Select Agents and Toxins (DSAT) at Centers for Disease Control and Prevention (Atlanta, GA), fulfilling all the requirements of an efficiently attenuated strain and thus not posing a threat to public health and safety (decision effective 12th December 2019 and available online
31. Centers for Disease Control Prevention. Biosafety in microbiological and biomedical laboratories (BMBL). St. Louis, MO: US Government Printing Office (2009).
58. Blodgett, R. Bacteriological analytical manual appendix 2: most probable number from serial dilutions,

Rongsheng Jin, biophysicist at the University of California, Irvine and lead author on the paper. He compares the complex to the toxin"s "landing gear".
This complex helps the toxin in two ways. First, as Jin implies, it lands on and then binds to the cells lining the inside of the intestine. "If they cannot stick there, [the toxin] would just go through and end up in the toilet," says Jin.
However, Jin says that understanding how these vehicles work could help scientists devise better ways to deliver drugs to the body. "We can make these proteins in the lab, assemble them into a vehicle and use them to deliver drugs to the intestine," he says.
Botulinum toxin (Botox) is a large protein that is composed of three domains (upper right corner, crystal structure shown in yellow, green, and pink). The new study reveals a novel mechanism by which the toxin hijacks three receptors on the host neuronal surface as its “GPS” — the peptide moiety of protein SV2 (green-blue ribbon model), a conserved glycan modification of SV2 (green and light blue sticks), and a lipid (brown sticks) — to launch its attack. Source: Rongsheng Jin and Guorui Yao / UCI
A study co-led by Rongsheng Jin, professor of physiology & biophysics at the University of California, Irvine; Min Dong with Boston Children’s Hospital-Harvard Medical School; and Andreas Rummel with the Hannover Medical School in Germany, reveals an important general mechanism by which the pathogen is attracted to, adapts to and takes advantage of glycan modifications in surface receptors to invade motor neurons. Glycans are chains of sugars synthesized by cells for their development, growth, functioning or survival. Results appear June 13 in Nature Structural and Molecular Biology.
“Our findings reveal a new paradigm of the everlasting host-pathogen arms race, where a pathogen develops a smart strategy to achieve highly specific binding to a host receptor while also tolerating genetic changes on the receptor,” Jin said. “And to some extent, this mechanism by which the toxin attacks human is similar to the one that is utilized by some important broad-neutralizing human antibodies to fight viruses, such as dengue viruses and HIV.”
Guorui Yao and Kwok-ho Lam at UCI, Sicai Zhang at Harvard, Stefan Mahrhold at the Hannover Medical School, Daniel Stern at Berlin’s Center for Biological Threats & Special Pathogens, Kay Perry at the Argonne National Laboratory in Illinois, and Karine Bagramyan and Markus Kalkum the Beckman Research Institute at the City of Hope in Duarte, Calif., contributed to the study, which was primarily supported by the National Institutes of Health.

2. The meeting registration is an integral package. We encourage all our participants to stay for the full meeting period and communicate with each other. No refund/discount or day pass is available for partial participation.
Hanwen Tong:Fudan University;Chenhao Xie:SenseDeal Intelligent Technology Co., Ltd.;Jiaqing Liang:Fudan University;Qianyu He:Fudan University;Zhiang Yue:Fudan University;Jingping Liu:East China University of Science and Technology;Yanghua Xiao:Fudan University,Fudan-Aishu Cognitive Intelligence Joint Research Center;Wenguang Wang:DataGrand Inc.
Yuqi Qin:Beijing University of Posts and Telecommunications;Pengfei Wang:Beijing University of Posts and Telecommunications;Biyu Ma:Xidian University;Zhe Zhang:Zhejiang University
Li Sun:North China Electric Power University;Junda Ye:Beijing University of Posts and Telecommunications;Hao Peng:Beihang University;Philip Yu:University of Illinois at Chicago
Hyunsik Jeon:Seoul National University;Jongjin Kim:Seoul National University;Hoyoung Yoon:Seoul National University;Jaeri Lee:Seoul National University;U Kang:Seoul National University
Haolun Wu:McGill University;Chen Ma:City University of Hong Kong;Yingxue Zhang:Huawei Noah"s Ark Lab;Xue Liu:McGill University;Ruiming Tang:Huawei Noah"s Ark Lab;Mark Coates:McGill University
Thanh Vinh Vo:National University of Singapore;Pengfei Wei:AI Lab Speech & Audio Bytedance;Trong Nghia Hoang:Washington State University;Tze Yun Leong:National University of Singapore
Yao Zhou:Instacart,University of Illinois Urbana Champaign;Jun Wu:University of Illinois Urbana Champaign;Haixun Wang:Instacart;Jingrui He:University of Illinois Urbana-Champaign
Hyeshin Chu:Ulsan National Institute of Science and Technology;Joohee Kim:Ulsan National Institute of Science and Technology;Seongouk Kim:Ulsan National Institute of Science and Technology;Hongkyu Lim:Ulsan National Institute of Science and Technology;Hyunwook Lee:Ulsan National Institute of Science and Technology;Seungmin Jin:Ulsan National Institute of Science and Technology;Jongeun Lee:Ulsan National Institute of Science and Technology;Taehwan Kim:Ulsan National Institute of Science and Technology;Sungahn Ko:Ulsan National Institute of Science and Technology
Zihan Liu:Zhejiang University,AI Lab, Westlake Institute for Advanced Study;Yun Luo:Westlake University;Lirong Wu:AI Lab, Westlake Institute for Advanced Study;Siyuan Li:AI Lab, Westlake Institute for Advanced Study;Zicheng Liu:AI Lab, Westlake Institute for Advanced Study;Stan Li:AI Lab, Westlake Institute for Advanced Study
Fuxian Li:Tsinghua University;Huan Yan:Tsinghua University;Guangyin Jin:Tsinghua University;Yue Liu:Alibaba Group;Yong Li:Tsinghua University;Depeng Jin:Tsinghua University
Ruitong Zhang:Beihang University;Hao Peng:Beihang University;Yingtong Dou:University of Illinois Chicago;Jia Wu:Macquarie University;Qingyun Sun:Beihang University;Yangyang Li:NERC-RPP, CAEIT;Jingyi Zhang:Beihang University;Philip S. Yu:University of Illinois Chicago
Xin Bi:Northeastern University;Shining Zhang:Northeastern University;Yu Zhang:Northeastern University;Lei Hu:Northeastern University;Wei Zhang:Northeastern University;Wenjing Niu:Northeastern University;Ye Yuan:Beijing Institute of Technology;Guoren Wang:Beijing Institute of Technology
Quanliang Jing:Institute of Computing Technology, Chinese Academy of Sciences,University of Chinese Academy of Sciences;Shuo Liu:Institute of Computing Technology, Chinese Academy of Sciences,University of Chinese Academy of Sciences;Xinxin Fan:Institute of Computing Technology, Chinese Academy of Sciences;Jingwei Li:Department of Computer Science and Engineering, University at Buffalo, SUNY;Di Yao:Institute of Computing Technology, Chinese Academy of Sciences;Baoli Wang:Microsoft Search Technology Center Asia;Jingping Bi:Institute of Computing Technology, Chinese Academy of Sciences
Jinwei Zeng:Tsinghua University;Guozhen Zhang:Tsinghua University;Can Rong:Tsinghua University;Jingtao Ding:Tsinghua University;Jian Yuan:Tsinghua University;Yong Li:Tsinghua University
Chen Wu:CAS Key Lab of Network Data Science and Technology, ICT, CAS,University of Chinese Academy of Sciences;Ruqing Zhang:CAS Key Lab of Network Data Science and Technology, ICT, CAS,University of Chinese Academy of Sciences;Jiafeng Guo:CAS Key Lab of Network Data Science and Technology, ICT, CAS,University of Chinese Academy of Sciences;Wei Chen:CAS Key Lab of Network Data Science and Technology, ICT, CAS,University of Chinese Academy of Sciences;Yixing Fan:CAS Key Lab of Network Data Science and Technology, ICT, CAS,University of Chinese Academy of Sciences;Maarten De Rijke:University of Amsterdam;Xueqi Cheng:Institute of Computing Technology, Chinese Academy of Sciences,University of Chinese Academy of Sciences
Bingning Wang:Tencent Inc.;Feiyang Lv:Tencent Inc;Ting Yao:Tencent Inc.;Yiming Yuan:Tencent Inc.;Jin Ma:Tencent Inc.;Yu Luo:Tencent Inc.;Haijin Liang:Tencent Inc.
Jiangxia Cao:Institute of Information Engineering, Chinese Academy of Sciences,School of Cyber Security, University of Chinese Academy of Sciences;Xin Cong:Institute of Information Engineering, Chinese Academy of Sciences,School of Cyber Security, University of Chinese Academy of Sciences;Jiawei Sheng:Institute of Information Engineering, Chinese Academy of Sciences,School of Cyber Security, University of Chinese Academy of Sciences;Tingwen Liu:Institute of Information Engineering, Chinese Academy of Sciences,School of Cyber Security, University of Chinese Academy of Sciences;Bin Wang:Xiaomi AI Lab, Xiaomi Inc.
Haoqi Zhang:Shanghai Jiao Tong University;Junqi Jin:Alibaba Group;Zhenzhe Zheng:Shanghai Jiao Tong University;Fan Wu:Shanghai Jiao Tong University;Haiyang Xu:Alibaba Group;Jian Xu:Alibaba Group
Jiangui Chen:CAS Key Lab of Network Data Science and Technology, ICT, CAS,University of Chinese Academy of Sciences;Ruqing Zhang:CAS Key Lab of Network Data Science and Technology, ICT, CAS,University of Chinese Academy of Sciences;Jiafeng Guo:CAS Key Lab of Network Data Science and Technology, ICT, CAS,University of Chinese Academy of Sciences;Yiqun Liu:BNRist, DCST, Tsinghua University;Yixing Fan:CAS Key Lab of Network Data Science and Technology, ICT, CAS,University of Chinese Academy of Sciences;Xueqi Cheng:CAS Key Lab of Network Data Science and Technology, ICT, CAS,University of Chinese Academy of Sciences
Phillip Howard:Intel Labs;Arden Ma:Intel Labs;Vasudev Lal:Intel Labs;Ana Paula Simoes:Intel Labs;Daniel Korat:Intel Labs;Oren Pereg:Intel Labs;Moshe Wasserblat:Intel Labs;Gadi Singer:Intel Labs
Kangzheng Liu:National Engineering Research Center for Big Data Technology and System, Services Computing Technology and System Lab, Cluster and Grid Computing Lab, School of Computer Science and Technology, Huazhong University of Science and Technology;Feng Zhao:National Engineering Research Center for Big Data Technology and System, Services Computing Technology and System Lab, Cluster and Grid Computing Lab, School of Computer Science and Technology, Huazhong University of Science and Technology;Hongxu Chen:Data Science and Machine Intelligence Lab, University of Technology Sydney;Yicong Li:Data Science and Machine Intelligence Lab, University of Technology Sydney;Guandong Xu:Data Science and Machine Intelligence Lab, University of Technology Sydney;Hai Jin:National Engineering Research Center for Big Data Technology and System, Services Computing Technology and System Lab, Cluster and Grid Computing Lab, School of Computer Science and Technology, Huazhong University of Science and Technology
Shuo Sun:Nanyang Technological University;Wanqi Xue:Nanyang Technological University;Rundong Wang:Nanyang Technological University;Xu He:Huawei Noah Ark Lab;Junlei Zhu:Webank;Jian Li:Tsinghua University;Bo An:Nanyang Technological University
Junwei Zhang:Tianjin University;Ruifang He:Tianjin University;Fengyu Guo:Tianjin Normal University;Jinsong Ma:Tianjin University;Mengnan Xiao:Tianjin University

"Now that we better understand the structure of the bacterial machinery that was designed for highly efficient toxin protection and delivery, we can see more clearly how to break it," said Rongsheng Jin, Ph.D., assistant professor in Sanford-Burnham"s Del E. Webb Neuroscience, Aging and Stem Cell Research Center and senior author of the study.
This latest study on the botulinum neurotoxin was the result of a close collaboration between the Jin group and a research group at the Institute of Toxicology at the Medical School of Hannover, led by Andreas Rummel, Ph.D., an expert on clostridial neurotoxins. They used a technique called X-ray crystallography, which uses powerful X-ray beams to produce 3D images of proteins at the atomic level, to study a genetically inactivated, nontoxic version of the botulinum neurotoxin.
"We were surprised to see that NTNHA, which is not toxic, turned out to be remarkably similar to botulinum neurotoxin. It"s composed of three parts, just like a copy of the toxin itself. These two proteins hug each other and interlock with what looks like a handshake," said Jin.
According to Jin, this new knowledge about how the botulinum neurotoxin and NTNHA balance the need for strong binding and a timely release could be exploited to outsmart them.
Moreover, this type of therapy could be designed for oral delivery, rather than injection, making it easier to treat large numbers of people during an outbreak. A similar strategy could be used to deliver other protein-based drugs that usually need to be injected. "Here, protein drugs could be linked to a botulinum neurotoxin fragment and protected with NTNHA. Then we could possibly take them by mouth," Jin said.




 8613371530291
8613371530291